In a previous post, we discussed how the methodology of "Clean In, Clean Through, Clean Out" provides a mechanism with which to deliver increased effectiveness of pathology services. In this post we will show how it has been used to study urinary tract infection and has resulted in delivery of more purpose, at less cost, with less harm and more capacity.
Trends in submission of urine specimens
The rate of submission of Mid Stream Urine samples (MSUs) from primary care has showed an inexorable year on year increase (fig.1). This is a pattern seen for many diagnostic tests.
fig 1. Number of urine samples received from primary care in North Devon 2002-2009. We looked at September only as it is the most stable month for requesting (no Bank Holidays etc)
What is the cause of this increase? Is it an ageing population? Or has clinical decision making changed? We do know that testing patients in whom it is not indicated is not benign.
Some consequences of sub-optimal urine microbiology
A patient harmed by unnecessary urine culture
JM was a well 67 year old woman who attended her surgery for a routine check up for hypertension. She was asked to bring a urine sample, which was dipped for protein. The specimen showed 1+ for leucocyte esterase and the healthcare assistant submitted the specimen to the laboratory for culture. No clinical details were entered on the request form. The laboratory isolated a pure growth of more than 10^8 organisms / litre of E coli which was resistant to trimethoprim and nitrofurantoin but sensitive to co-amoxiclav and ciprofloxacin, and this was reported to the GP. The GP had never seen the patient, but presumed that she must have a urinary tract infection on the basis of this result (although there was no clinical evidence of infection). As she was allergic to penicillin, the GP prescribed ciprofloxacin. One week later, JM returned to see her GP with severe diarrhoea. A sample was sent to the laboratory and Clostridium difficile was detected.
Unnecessary antibiotics given because of unnecessary urine culture
We looked at 40 patients from primary care with a pure growth of E. coli. 30 of these patients had symptoms of infection and were already on antibiotic treatment. 10 patients had no symptoms of infection, but had had samples sent for just the reasons described above (ie. positive dipsticks on routine screening for other conditions). All these patients were prescribed antibiotics as a result of the test. This is clear harm to the patient driven by inappropriate testing.
It is also worth considering the effect of abnormal results on the GP. The abnormal result will need to be considered when it arrives back. The choices are to ignore it or to act. Ignoring a test is not trivial, and comes with an opportunity cost - there has to be a conscious decision not to act, or there is a danger that abnormal results performed for good reasons may be erroneously ignored. This is an ever increasing problem with the inexorable rise of testing. And as we have seen the decision to act can be even more problematic - not only does it take time to 'do something', there is a very real risk that this action will lead either to direct harm to the patient (eg. antibiotic prescribing), or more insidiously perpetuate further work in the system ('failure demand') - bad for the patient and bad for the sustainability of healthcare.
We applied our definitions of points of leverage to study and act upon the problem.
Improving Clean In for urine microbiology
1. Were tests necessary?
We looked at the request details of individual specimens being sent to the laboratory. We saw a large number of requests did not include important relevant clinical information, but instead were either blank, or contained information pertaining to a urinary dipstick (fig 2). It is worth noting that urinary tract infection is generally a clinical diagnosis that the laboratory investigations can only support - so it is hard to know what "?UTI" means.
fig. 2 Clinical details on request forms submitted with urine specimens from primary care 2010.
We asked why these tests were being sent and to do this went to individual case records in primary care. What we saw is that tests were being ordered by health care assistants who were seeing patients for chronic disease reviews and that much of the new demand appears related to the introduction of primary care quality targets for monitoring of chronic kidney disease.
Assessment of kidney damage in these patients requires an assessment of the amount of protein excreted in the urine. A quick way of doing this is to use point of care urine dipsticks. Many dipsticks provide multiple additional tests - irrelevant in the management of chronic kidney disease, but of possible utility in other situations, such as determining likelihood of urinary tract infection in patients with vague symptoms of this condition.
And here we see the rub - the dangers of testing that is not aligned to a clinical question. These patients with chronic kidney disease who are presenting for routine 'check ups' (and so in whom we would not be worrying about infection) often give positive dipstick results for nitrites and leucocyte esterase - which are associated with infection. However, dipsticks have a high sensitivity but a poor specificity for diagnosing infection (ie. a negative test is good at ruling out infection, but a positive test does not diagnose infection). But, perhaps not wanting to ‘miss something important’, the urine sample is sent to the laboratory to look for bacterial growth - traditionally seen as the gold standard arbiter of infection.
There is now a second misunderstanding related to the gold standard nature of urine culture (interestingly perpetuated in much of the literature on urine infection). Clinicians are often taught that a growth of 10^8 organisms per litre is diagnostic of infection. The original work by Kass on this subject did not claim this - rather it showed that a pure growth of this magnitude was predictive of reproducibility of this growth in a research setting. This is most definitely not the same as a diagnosis of infection in a routine clinic setting. The incidence of asymptomatic colonisation of the urinary tract is high - and there is good evidence that this should not be treated with antibiotics, except in well defined populations (eg. pregnant women). In addition, the quality of urine specimens is often poor from routine settings, with samples frequently contaminated with perineal flora. There then may be substantial delay getting these specimens to the laboratory, and specimen stabilisation is never absolute. The consequence of all this is that the performance of urine culture in routine practice is significantly worse than that seen in the research setting. To give some idea of the magnitude of this problem, all newly pregnant women are asked to send a urine specimen for culture (as this is a group in whom asymptomatic bacteriuria is a very high risk for causing pyelonephritis). Our laboratory requests repeat samples from all pregnant women if they have no symptoms but a pure growth of organisms. We find that this pure growth can only be reproduced in one third of patients. In other words, the specificity of a pure growth of bacteria for diagnosing UTI is very poor. As with all tests, the prior probability of the condition being present is key when trying to interpret a test result, be it the result of a point of care test (eg. dipstick) or a laboratory test (eg. culture).
Acting on “Clean In” as a point of leverage for improving the use of urine culture
It is easy to see now that acting upon the “Necessary” component of Clean In is a key point of leverage for improving the effectiveness of urine culture as part of the informed management of urinary tract infection.
To do this, we went to the source of the problem. We talked to Health Care Assistants (HCAs) about the way they work and what would help them to be better. We heard that they often work with guidelines that are partially conflicting. We heard how HCAs take great pride in what they do and are keen to ‘do the right thing’. We heard how noone had talked to them about possible harms of inappropriate testing.
To address these issues we worked with HCAs to design algorithms for the management of urine dipsticking. We came up with simple visual cues that distinguish between specimens that are for diagnosis of infection (red top tubes containing borate for bacterial stabilisation) and specimens that are for diagnosis of kidney damage (white top tubes). Our algorithms resolved or clarified areas of advice that could have been seen as in conflict.
We introduced this guideline into a pilot practice and saw an immediate and dramatic effect on urine culture submissions from 140 per month to just 7 per month. We were worried that a consequence of our advice had been that clinicians were not sending urine for culture in patients with infection. In other words, it is possible we had had a negative impact on the “Maximally Appropriate” aspect of Clean In. In an effort to improve the “Sufficient”and “Maximally appropriate” aspects of urine submission, we promoted national guidance on the management of urinary tract infection, which includes advice on when to submit specimens for laboratory analysis. We saw specimen submissions rise again in the pilot practice to around 50% of the previous peak and the quality of information provided also improved.
With this clear success, it was easy to roll out the programme across North Devon. We promoted the dipsticking algorithm, the guidance on management of urinary tract infection and a distillation of our finding as a series of “Top Tips”. We visited all practices and spoke to doctors and HCAs. We gave presentations at regional education days. As a consequence of this work, we have seen a sustained reduction in urine specimens submitted for culture from primary care (fig 3) and an increase in the quality of the information provided (fig 4).
fig 3. Number of urine samples received from primary care in North Devon 2002-2015
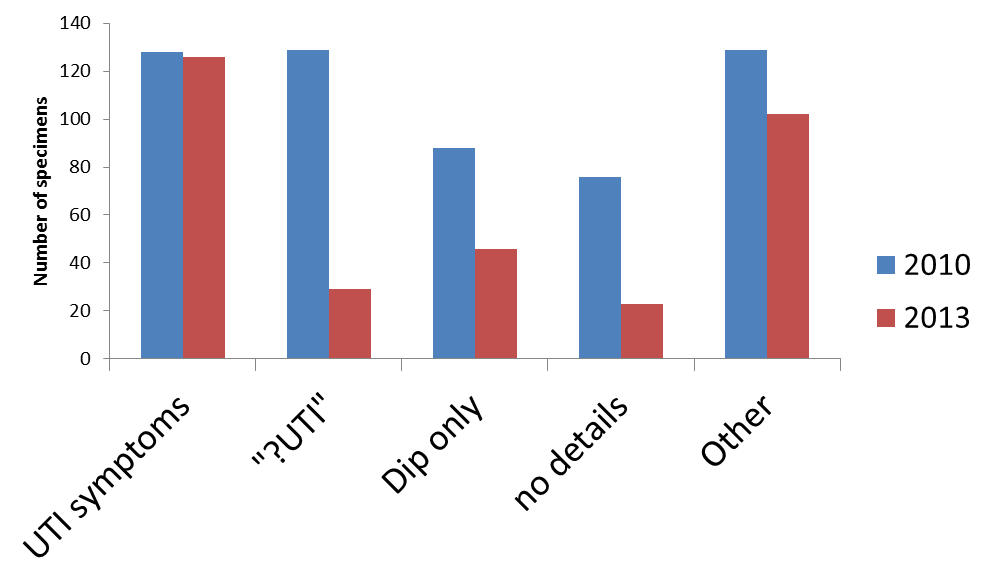
fig 4. Clinical details submitted with urine specimens from primary care
Clean Through - Using improvements from Clean In to improve Specimen processing
As a result of our work improving Clean In we saw a number of benefits in the laboratory. First, we had more time to redesign processes against purpose as we were doing less work (which had no purpose). Second, because we now knew that specimens were likely to be of clinical importance, we could easily see the value the laboratory was contributing to the management of these patients, and ask ourselves “Are we doing all we can to help?”. “Clean Through” has proved to be a useful framework.
What does it mean to process a urine specimen without error?
Laboratories have many ways of controlling processes to reduce error. Some of these are picked up as part of the accreditation process under ISO15189. How do we explain this to clinicians and patients in a way that they would find reassuring? Internal and External Quality Assurance (IQA and EQA) should be designed around answering this question. It is reassuring to know that a lab has performed well in an EQA process that is designed to test its ability to analyse accurately specimens that are relevant to a specific case. This could, in principle, form part of a pathology report. For instance, we would be in the position to add a comment for urine microbiology that states our confidence in our ability to identify correctly an organism, and to report accurately a set of antibiotic sensitivities.
EQA schemes are important ways to test and challenge performance against known standards. However, these are readily identifiable specimens and an accurate result does not necessarily imply that this would be achieved in a real life clinical context, To answer this question, we need a well designed IQA process. For urine microbiology. we think this must involve running specimens in parallel, ideally in a way that means they are blinded to all people involved in the process. If this is built into the routine working environment then we can easily spot variation.
However, microbiology is a dirty science. It is hard to get perfect specimens in a real life clinical setting, and microorganisms do not always behave in an entirely predictable way. As an example in urine microbiology, a substantial number of specimens contain mixtures of bacteria. Traditional teaching is that this represents contamination and that the samples should be rejected. However, when we look at patients who are bacteraemic from urine infections (in other words the bacteria have spilt over from the urine into the blood) we see that about 20% of patients have a concurrent urine that has mixtures of organisms in it. When we look at this mixture, we can see important clinical information that can be elucidated quickly without little work. On direct antibiotic sensitivity testing, it is obvious to the scientist reading the plates which antibiotics are likely to be good bets, and which should be avoided (fig 5).
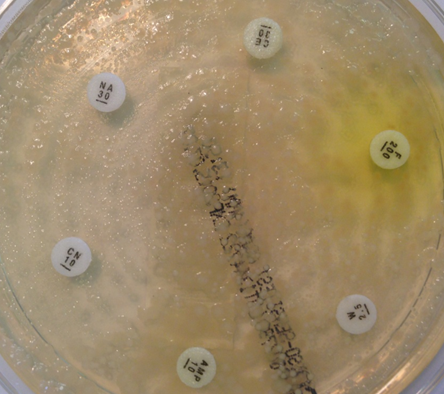

Fig 5. Reading mixed sensitivities. This specimen from a patient with urinary sepsis contains at least 3 organisms. Some, but not all, look resistant to gentamicin, so we would use this antibiotic with caution. Most organisms look resistant to amoxicillin, so we would avoid this antibiotic. All organisms look sensitive to meropenem, and this would be a good choice if the patient had ongoing sepsis.
The use of mixed culture sensitivity in a patient with severe urinary sepsis
GB was a 82 year old man with a long term urinary catheter due to urinary retention. He was admitted to hospital with a high fever and low blood pressure. A diagnosis of urinary tract infection was made and blood cultures and a catheter urine specimen were sent to the laboratory. The patient was given amoxicillin plus gentamicin and resuscitated. The following day the urine plates had at least 3 different organisms on them, at least one of which was resistant to gentamicin. All isolates looked sensitive to ciprofloxacin. The patient was reviewed by the consultant microbiologist on the medical assessment unit. He still had a high fever and had an episode of rigors during the night. His blood pressure had stabilised but he looked unwell. The patient was switched to ciprofloxacin. Later that day his blood cultures became positive with a gram negative organism. This turned out to be a gentamicin resistant, ciprofloxacin sensitive organism. The patient made an uneventful recovery and was discharged 2 days later after a catheter change.
Error defined in a way that is relevant to patient care
The trouble for laboratories is that it is hard to standardise these processes, and under rigorous scientific conditions it would be appropriate to reject the specimen and start again. The trouble for the patient and their carers is that infection is a time limited condition - they do not have days in which to get the perfect result. What is needed is a result that is clinically helpful in a timeframe that is clinically relevant. Laboratories have a choice when things do not behave perfectly. They can reject them, or analyse them in great detail. The first of these does little to help the patient, and the second takes time and is likely to be costly. A third way is to explore ways to report mixed cultures in a way that is helpful, without overstating the certainty of the result and the potential for error. In our laboratory, we have designed new methods that allow the laboratory scientists to convey what they see on the plates to the consultant microbiologist on the preliminary report. The consultant then takes this information and adds a comment on therapeutic options based on the clinical details given by the requesting clinician. We will see the benefit of this approach in terms of reporting and timeliness below.
What does it mean to process a specimen with known variation?
Variation in diagnostics is well recognised, and yet is rarely considered when results are applied to clinical practice. Even microbiologists will see an antibiotic sensitivity result in a report and treat it as incontrovertible truth. So how confident should we be in a result? What is the variation that is intrinsic to the analytical process? What is the variation that is extrinsic to the analytical process (including, for instance, variations in expression of resistance genes or host excretion of antimicrobial factors)? Laboratories are most concerned with minimising intrinsic variation, but to a patient and their carer all uncertainty is common.
We have started to use our IQA processes to examine uncertainty in antibiotic sensitivity testing. We process test specimens in duplicate and see if we get the same result. Sometimes we see special cause errors (for example, an E coli reported as a Proteus) and this leads us to examine how they can be prevented (see above). But other times we see common cause, predictable variation in results. About 10% of antibiotic sensitivities can change between sensitive and resistant. This is a somewhat surprising result at first pass. But when we examine the science behind sensitivity testing we should not be too surprised. In controlled settings, antibiotic zone sizes can vary by up to 10%. When an organism can change between sensitive and resistant based on zone size differences of just 1mm, it is no surprise that there is variation. This begs the question of what this means at a clinical level. As a clinician, how do I interpret sensitivity results? This is difficult terrain, but it must start with an honest dialogue about what our results really imply.
What is ‘On time’ for urine microbiology?
The issue of timeliness depends to some extent on the question being asked, If we are asking “Does this pregnant woman have asymptomatic bacteriuria?” then there is little need to issue a result within 48 hours. If we are asking, “Does this patient with severe urinary sepsis have an organism resistant to the antibiotics I am going to give?” then the timeframe is clearly much shorter. In an ideal world, we would want a result there and then. But given that it is possible to have preliminary sensitivities from a well inoculated agar plate within a few hours, then we should ask perhaps why this is not done more frequently. A more common question in primary care is “This patient has pyelonephritis. I am going to treat them with empirical antibiotics, and then review them tomorrow. If they are not better, I want to know if it is because I have chosen an antibiotic to which the organism is resistant, and if so, what would be a better choice?” In order for this to happen, the specimen must arrive in the laboratory on the day it is taken and be plated up with direct sensitivities. It must be read, interpreted and validated the next day.
In North Devon, one of the consequences of improving Clean In was a substantial reduction in workload that meant we were able to experiment with ways of improving the number of results released the day after specimen receipt. We looked at the reasons we were not releasing results the following day using a standard panel of six antibiotics for direct sensitivity testing. We found that predictably it was because of coliforms resistant to first line antibiotics, or Pseudomonas, or Streptococci. We showed that by using 12 antibiotic discs on two plates we could get all the sensitivity and organism identification data that was required to release a report. We also showed that manual microscopy was faster than an expensive automated method for detecting pyuria, and we saved £20000 by removing this equipment (which was substantially more than the cost of the additional agar plates that were required). As described above, we also developed ways for dealing quickly and appropriately with mixed cultures. We also changed laboratory working practices so that specimens received before 10pm were inoculated, and plates were read every day, including Sundays and Bank Holidays. The result of these changes on our ability to release next day sensitivity results is shown in fig. 6
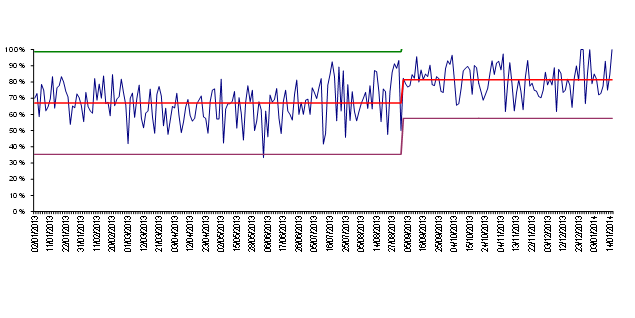
Fig 6. Percent of urine specimens released on the day after receipt
Clean Out - Changing the way results are reported
We are at the beginning of exploring how we act upon the points of leverage identified here. We know that results are often misinterpreted by clinicians, and that this happens in many ways. We know that antibiotics are started when they are not indicated, just because organisms have been isolated; we know that antibiotics are switched on the back of resistance that is not clinically relevant; we know that mixed cultures are interpreted as contamination when the patient has clinical evidence of urinary tract infection. These are complex areas to address.
Making results more helpful
One approach to the problem is to realise that culture and sensitivity reports are often overly simplistic ways of answering the clinical question that has been posed. We have begun to add more consultant narrative to reports in a bid to aid the clinical decision (fig 7).
Fig 7.Percent of urine reports with consultant microbiologist advice
We also see education about management of infection as a key part of Clean Out. Much infection management is based on empirical clinical decision making, and clinicians have to make complex decisions about which antibiotics to give. This must take into account the local and patient-specific antibiotic resistance patterns. It should also consider the potential for harm, including that caused by broad-spectrum antibiotics. We ensure that our laboratory results are matched to this advice. In order to do this, we have produced comprehensive treatment decision aids for clinicians, and back this up with frequent and varied educational events.
Helping results to be understood
We are often told that patients ‘want’ antibiotics, but when we talk to patients we find the truth is more nuanced than this. They really want reassurance and appropriate treatments, they do not want to suffer needless side effects. Giving patients reassurance is, again, a complex area. We work with social marketing teams in public health departments to design messages that speak to patients and help them make informed decisions, such as the Listen To Your Gut campaign. We now see how our approach of Clean In, Through, and Out complements, or even fulfils, the needs of an antibiotic stewardship programme.
Results that reflect what is normal for me, not normal for the population
MV is a 57 year old female who has suffered with recurrent urinary tract infections since undergoing an enterocystoplasty for interstitial cystitis. These infections were severe enough to prevent her from working. Despite numerous attempts to eradicate these infections (including quasi-experimental treatments such as high dose prolonged combination antibiotics; hyaluronic acid; intravesicular gentamicin) we have been unable to stop breakthrough infections, while inflicting a wide range of serious side effects, from anaphylaxis, to tendon rupture and ototoxicity. The organisms we isolate at each episode are unpredictable, with unpredictable antibiotic sensitivities, and in other patients would often be dismissed as contaminants (including, frequently, non-aeruginosa Pseudomonas and non-haemolytic streptococci). Working with the patient, we have discovered that she is aware when an infection is imminent, and can submit a specimen directly to the laboratory. We are able to plate this as soon as it arrives, and can give preliminary organism identification and sensitivity data within 6 hours. This allows early, rational antibiotic initiation. We have shown that this has reduced the duration and severity of her infections, and that her total antibiotic exposure is markedly reduced. She has returned to work and is able to play tennis again.
We know that there is a significant group of patients, like MV, with complex infections that are ill served by current services. What is ‘normal for the population’ may not be ‘normal for me’. This group of patients do not follow the accepted paradigms and require non-standard approaches. We are increasingly using the laboratory in flexible ways to support early and tailored treatment regimes. Although these are isolated cases, they serve to remind us of our purpose. We are not here to generate results, we are here to help patients and their carers make informed decisions. Sometimes this will be easy, sometimes we will be wrong, but at all times this purpose acts as a guiding principle by which we are willing to be judged.
The impact of focussing on points of leverage on system measures
As we have discussed, these changes have led to clear improvements in capacity of the service to do purposeful work. We have seen significant benefits to patient care at an individual level. Is it possible to see benefits in aggregate data? Unfortunately, the data is often poor, and open to bias. But we can see a number of things falling out of this work :
Cost to commissioners has reduced by approximately £200 000 per annum
Mortality following gram negative bacteraemia is the lowest in the region (6.1% seven day mortality; vs 7.1% and 9.0% in neighbouring hospitals).
Use of antibiotics in North Devon for urinary tract infection has fallen by 24%
Carbapenem prescribing has fallen by approximately 50%
Admission rates for urinary tract infection have stabilised having being rising at 10-20% per annum.
We have been able to absorb the loss of a biomedical scientist without replacement.
Consumable costs have fallen by approximately £20000 per annum
Conclusions
Understanding of purpose takes us out of the laboratory and out of the transactional context of the request for testing that the laboratory receives. It takes us to where citizens are and to what difference they want us to help them make in their lives. When we do this we see different and better problems and find different and better solutions. The points of intervention are often not where we traditionally focus in a narrow framed approach. By identifying simple points of leverage, and by looking from the outside-in (ie from the perspective of the patient, and considering our purpose), we see our work as part of a wider system and have a mechanism for solving the problems that matter to citizens. Any other approach (eg. a focus on targets; thinking from the inside out) risks solving the wrong problems.
Acting at these points of leverage means better outcomes for the citizen, more purposeful work for those working in the system, less demand into the system and therefore more capacity to do even more good things. This creates a virtuous cycle: taking the time to understand and do what is really necessary, creates more time to understand and do what is really necessary.
In subsequent posts, we will see how this method has been applied to patients with chronic wounds, and patients with chronic diseases.
